| CRM: |
| CE8012 |
|---|
| Overlaps: |
| pdm2 neurogenic |
|---|
| Reference: |
| Berman, 2004 |
|---|
| Berman 2002 ID: |
| 5 |
|---|
| Crm Activity: |
| + |
|---|
| 5' Primer: |
| 5'-TTAGGCGCGCCGGAGTCGGTTGGTGTAGCG-3' |
|---|
| 3' Primer: |
| 5'-ATTGCGGCCGCGGGTTTATGGGCATTAGTTGG-3' |
|---|
|
| D.mel fasta: |
| dmel unflanked |
|---|
| pCRM region: |
| genomic region |
|---|
| Chrom arm: |
| 2L |
|---|
| pCRM start: |
| 12,663,605 |
|---|
| pCRM end: |
| 12,664,915 |
|---|
| pCRM len: |
| 1,311 |
|---|
|
| D.pse region homologous: |
| 1 |
|---|
| D.pse fasta: |
| dpse unflanked |
|---|
| dpse arm: |
| Dpse_3212646_3212123_3213189_1_68286 |
|---|
| dpse start: |
| 27,914 |
|---|
| dpse end: |
| 29,845 |
|---|
| dpse len: |
| 1,932 |
|---|
|
| Flanked D.mel fasta: |
| dmel flanked |
|---|
| Flanked D.pse fasta: |
| dpse flanked |
|---|
| LAGAN alignment (readable): |
| alignment |
|---|
| LAGAN alignment (mfa): |
| alignment |
|---|
|
| D.mel binding site xml: |
| xml |
|---|
|
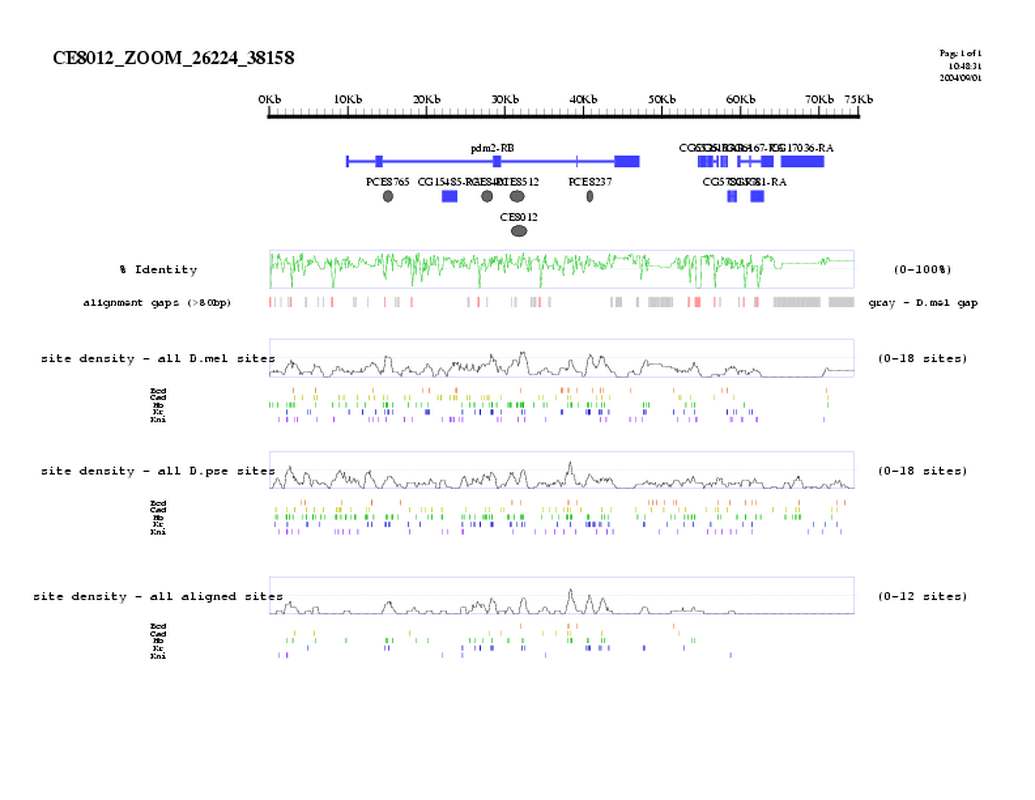
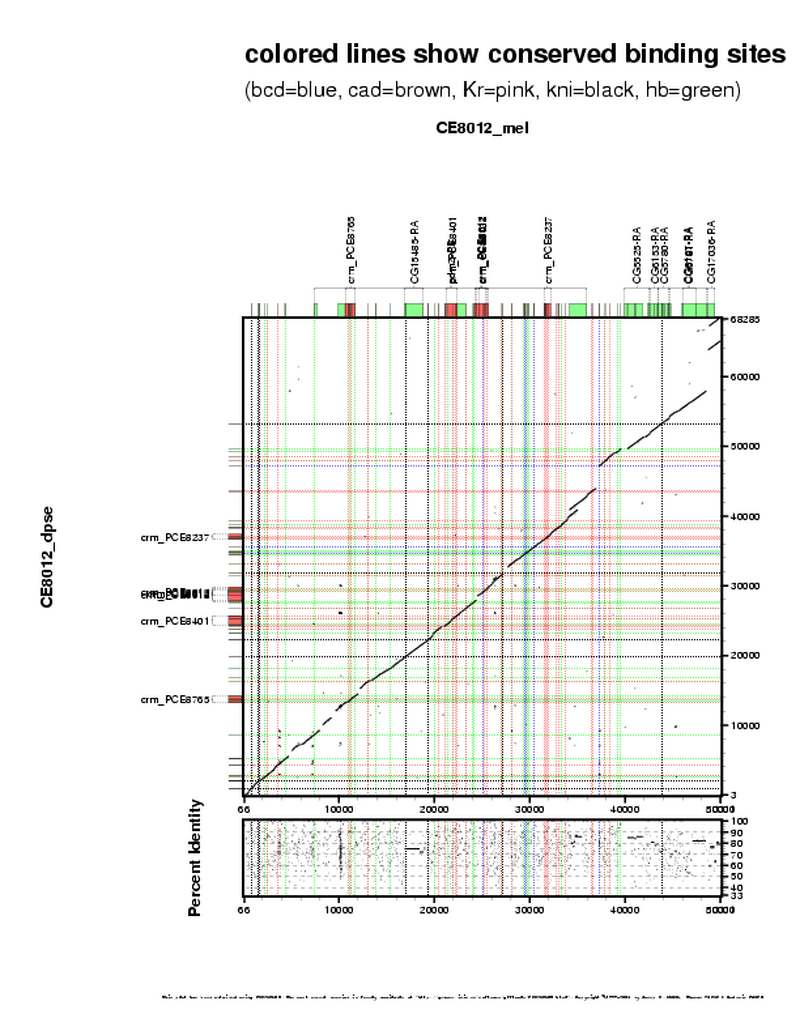
| overall site conservation z-score: |
| 4.0 |
|---|
|
| Percent identity: |
| 69.6% |
|---|
|
| D.mel sites: |
| 16 |
|---|
| D.pse sites: |
| 15 |
|---|
| Aligned sites: |
| 5 |
|---|
| Aligned + preserved sites: |
| 12 |
|---|
|
| D.mel site dens: |
| 12.2 |
|---|
| D.pse site dens: |
| 7.8 |
|---|
| Aligned site dens: |
| 3.8 |
|---|
| Aligned + preserved site dens: |
| 9.2 |
|---|
|
| pCRM position relative to 5' gene: |
| upstream |
|---|
| pCRM position 5' gene relative: |
| -5,566 |
|---|
| 5' gene CG: |
| CG15485 |
|---|
| 5' gene: |
| CG15485 |
|---|
| 5' gene insitu: |
| CG15485 insitu |
|---|
| 5' gene flybase: |
| CG15485 flybase |
|---|
| 5' gene start: |
| 12,658,039 |
|---|
| 5' gene end: |
| 12,656,134 |
|---|
| 5' gene txl start: |
| 12,658,039 |
|---|
|
| pCRM position 3' gene relative: |
| within_intron |
|---|
| pCRM position relative to 3' gene: |
| 16,907 |
|---|
| 3' gene CG: |
| CG12287 |
|---|
| 3' gene: |
| pdm2 |
|---|
| 3' gene insitu: |
| pdm2 insitu |
|---|
| 3' gene flybase: |
| pdm2 flybase |
|---|
| 3' gene start: |
| 12,646,698 |
|---|
| 3' gene end: |
| 12,675,183 |
|---|
| 3' gene txl start: |
| 12,649,335 |
|---|